QUEFTS¶
Introduction¶
This chapter describes the QUEFTS model. QUEFTS stands for
QUantitative Evaluation of the Fertility of Tropical Soils. It is a
simple rule-based model to estimate crop yield given the amount of
fertilizer applied, or to estimate the amount of fertilizer needed to
reach a particular yield.
To run the model, you need soil and crop parameters, as well as the amount of fertilizer applied, and the attainable crop biomass production. In this context, the attainable production that is either the maximum production in the absence of nutrient limitation, or another (lower) target biomass production.
QUEFTS was first described in: B.H. Janssen, F.C.T. Guiking, D. van der Eijk, E.M.A. Smaling, J. Wolf and H.van Reuler. 1990. A system for quantitative evaluation of the fertility of tropical soils (QUEFTS). Geoderma 46: 299-318.
Requirements¶
We will use the R package Rquefts. You can install it from CRAN.
if (!require(Rquefts)) { install.packages("Rquefts") }
library(Rquefts)
Technical background¶
Potential supply of nutrients¶
The model uses soil properties to estimate the potential soil supply of nitrogen (N), phosphorus (P) and potassium (K). This is done based on relationships between chemical properties of the topsoil and the maximum quantity of those nutrients that can be taken up by the crop, if no other nutrients limiting growth. This amount is crop and soil specific and can be determined through experiments where all other nutrients are in ample supply. The target nutrient thus becomes the most limiting, and the maximum soil supply of that nutrient can then be estimated from the crop’s nutrient uptake, and related to soil properties such as pH, and organic carbon content (Janssen et al., 1990). The equations used by QUEFTS to calculate the potential supplies of nutrients from soil properties were derived from empirical data of field trials in Suriname and Kenya. They are thought to be applicable to well drained, deep soils, that have a pH(H20) in the range 4.5-7.0, and values for organic carbon below 70 g/kg, P-Olsen below 30 mg/kg and exchangeable potassium below 30 mmol/kg.
Let’s have a look at this relationship. Below we compute the nutrient supply for a range of pH values, and given different levels for the other chemical properties.
library(Rquefts)
# range of pH values
ph <- seq(4, 8, 1)
s <- nutSupply(ph, SOC=1.5, Kex=1, Polsen=1)
cbind(ph, s)
## ph N_base_supply P_base_supply K_base_supply
## [1,] 4 2.55 -0.0250 134.32836
## [2,] 5 5.10 0.7625 104.47761
## [3,] 6 7.65 1.0250 74.62687
## [4,] 7 10.20 0.7625 44.77612
## [5,] 8 12.75 -0.0250 14.92537
With a more detailed range of pH values, to make plots.
ph <- seq(4, 8, 0.25)
# three combinations
s1 <- nutSupply(ph, SOC=15, Kex=5, Polsen=5)
s2 <- nutSupply(ph, SOC=30, Kex=10, Polsen=10)
s3 <- nutSupply(ph, SOC=45, Kex=20, Polsen=20)
Now plots of the effect of pH on N, P and K supply, for three situations (low, medium and high levels of organic matter, exchangeble K, and Olsen-P).
par(mfrow=c(1,3))
for (supply in c("N_base_supply", "P_base_supply", "K_base_supply")) {
plot(ph, s3[,supply], xlab="pH", ylab=paste(supply, "(kg/ha)"), type="l", ylim=c(0,max(s3[,supply])))
lines(ph, s2[,supply])
lines(ph, s1[,supply])
}
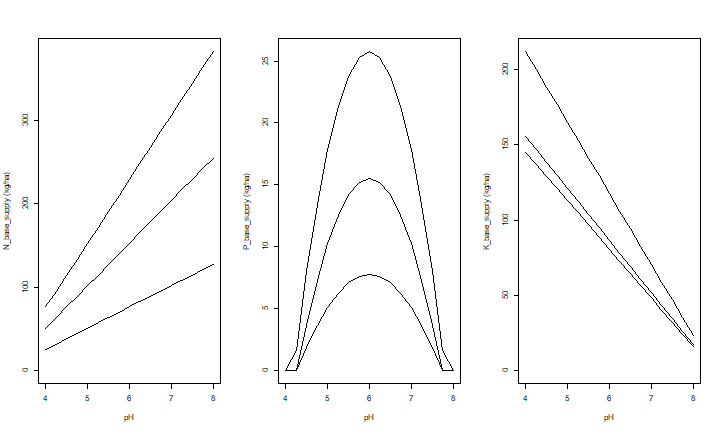
This shows that P-supply is highest at a pH=6, for any Olsen-P level. N-supply decreases with pH (given the amount of soil Carbon), and K-supply increases with pH (given the amount of exchangeble K).
Now let’s look at nutrient supply for varying levels of Soil Carbon.
# unit is g/kg
om <- seq(0, 70, 5)
# three combinations
s1 <- nutSupply(pH=5.5, SOC=om, Kex=5, Polsen=5)
s2 <- nutSupply(pH=5.5, SOC=om, Kex=10, Polsen=10)
s3 <- nutSupply(pH=5.5, SOC=om, Kex=20, Polsen=20)
# and plot
par(mfrow=c(1,3))
for (supply in c("N_base_supply", "P_base_supply", "K_base_supply")) {
plot(om, s3[,supply], xlab="Soil Carbon (g/kg)", ylab=paste(supply, "(kg/ha)"), type="l", ylim=c(0,max(s3[,supply])))
lines(om, s2[,supply])
lines(om, s1[,supply])
}
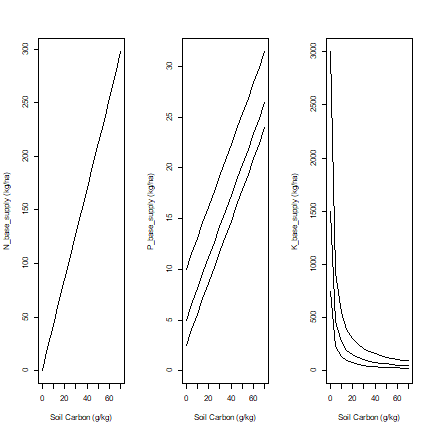
This shows that organic carbon has a strong positive effect on available N and P, but a negative effect on K availability (given the amount of exchangable K).
We can assume that these relationships are robust, at least for the conditions that they were designed for (pH(H20) 4.5-7.0, organic carbon below 70 g/kg, P-Olsen below 30 mg/kg and exchangeable potassium below 30 mmol/kg). But with more data, they could be re-estimated, also for other conditions.
Actual nutrient uptake¶
The actual uptake of each nutrient is estimated as a function of the potential soil supply of that nutrient, taking into account the potential soil supply of the other two nutrients, and the maximum nutrient concentrations in the vegetative and generative organs of the crop.
The estimated uptake of nitrogen, phosphorus, and potassium is used to estimated biomass production ranges for each nutrient. For each pair of nutrients two yield estimates are calculated. For example the yield for the actual N uptake is computed dependent on the yield range for the P uptake, and that for the actual P uptake in dependent on the yield range for the N uptake. This leads to six combinations describing the uptake of one nutrient given maximum dilution or accumulation of another.
The nutrient-limited yield is then estimated by averaging the six yield estimates for paired nutrients. An estimate based on two nutrients may not exceed the upper limit of the yield range of the third nutrient, that is, the concentration of a nutrient can not be lower than its maximum dilution level. Neither may the yield estimates exceed the spacified maximum (attainable) production.
TO DO: illustrate the procedure numerically and graphically
TO DO: discuss limitations based on the Sattari et al paper
Create and run a model¶
The first step is to create a model. You can create a model with default parameters like this
library(Rquefts)
q <- quefts()
And then run the model
run(q)
## leaf_lim stem_lim store_lim N_supply P_supply K_supply
## 1928.19667 1928.19667 3756.94929 90.00000 11.00000 90.00000
## N_uptake P_uptake K_uptake N_gap P_gap K_gap
## 81.66872 10.88951 77.38765 60.00000 205.00000 0.00000
You can also initialize a model with parameters of you choice
soiltype <- quefts_soil()
barley <- quefts_crop('Barley')
fertilizer <- list(N=50, P=0, K=0)
biomass <- list(leaf_att=2200, stem_att=2700, store_att=4800, SeasonLength=110)
q <- quefts(soiltype, barley, fertilizer, biomass)
TODO discuss parameters
If the crop has biological nitrogen fixation (in symbiosis with Rhizobium) you can use the NFIX parameter to determine the fraction of the total nitrogen supply that is supplied by biological fixation. Note that NFIX is a constant, while in reality nitrogen fixation is amongst other things dependent on the amount of mineral nitrogen available in the soil.
Explore¶
library(Rquefts)
rice <- quefts_crop("Rice")
q <- quefts(crop=rice)
q$leaf_att <- 2651
q$stem_att <- 5053
q$store_att <- 8208
fert(q) <- list(N=0, P=0, K=0)
N <- seq(1, 200, 10)
results <- matrix(nrow=length(N), ncol=12)
colnames(results) <- names(run(q))
for (i in 1:length(N)) {
q["N"] <- N[i]
results[i,] <- run(q)
}
yield <- results[,"store_lim"]
par(mfrow=c(1,2))
plot(N, yield, type="l", lwd=2)
Efficiency = yield / N
plot(N, Efficiency, type="l", lwd=2)

f <- expand.grid(N=seq(0,400,10), P=seq(0,400,10), K=c(0,200))
x <- rep(NA, nrow(f))
for (i in 1:nrow(f)) {0
q$N <- f$N[i]
q$P <- f$P[i]
q$K <- f$K[i]
x[i] <- run(q)["store_lim"]
}
x <- cbind(f, x)
# display results
library(raster )
r0 <- rasterFromXYZ(x[x$K==0, -3])
r200 <- rasterFromXYZ(x[x$K==200, -3])
par(mfrow=c(1,2))
plot(r0, xlab="N fertilizer", ylab="P fertilizer", las=1, main="K=0")
contour(r0, add=TRUE)
plot(r200, xlab="N fertilizer", ylab="P fertilizer", las=1, main="K=200")
contour(r200, add=TRUE)
#plot(stack(r0, r200))
Callibration¶
Here I illustrate some basic approaches that can be used for callibration. First I generate some data.
set.seed(99)
yldfun <- function(N, noise=TRUE) { 1000 + 500* log(N+1)/3 + noise * runif(length(N), -500, 500) }
N <- seq(0,300,25)
Y <- replicate(10, yldfun(N))
obs <- cbind(N, Y)
We will use Root Mean Square Error (RMSE) to assess model fit.
RMSE <- function(obs, pred) sqrt(mean((obs - pred)^2))
Let’s see how good the default parameters work for us.
q <- quefts()
q$P <- 0
q$K <- 0
results <- matrix(nrow=length(N), ncol=12)
colnames(results) <- names(run(q))
for (i in 1:length(N)) {
q$N <- N[i]
results[i,] <- run(q)
}
yield <- results[,'store_lim']
RMSE(obs[,-1], yield)
## [1] 1868.019
The RMSE is quite high.
Create a function to minimize with the optimizer. Here I try to improve the model by altering four parameters.
vars <- c('soil$N_base_supply', 'soil$N_recovery', 'crop$NminStore', 'crop$NmaxStore')
f <- function(p) {
if (any (p < 0)) return(Inf)
if (p['crop$NminStore'] >= p['crop$NmaxStore']) return(Inf)
if (p['soil$N_recovery'] > .6 | p['soil$N_recovery'] < .3) return(Inf)
q[names(p)] <- p
pred <- run(q)
RMSE(obs[,-1], pred['store_lim'])
}
# try the function with some initial values
x <- c(50, 0.5, 0.011, 0.035)
names(x) <- vars
f(x)
## [1] 1573.818
Now optimize
par <- optim(x, f)
# RMSE
par$value
## [1] 1478.384
# optimal parameters
par$par
## soil$N_base_supply soil$N_recovery crop$NminStore
## 0.01398939 0.59768399 26.37808069
## crop$NmaxStore
## 37.05140516